Downloading Data from UK Biobank
1. Register as a Researcher
- Go to the UK Biobank Access Management System (AMS): ams.ukbiobank.ac.uk
- Create an account using your email - can be personal or Rutgers.
- Send Avram the email you used to sign up, and ask him to add you to as a collaborator to our lab project, “Structural genetic contributions brain function and behavior”, application ID 25163
Once you’re added, check your account profile to see if you’ve been added to a project. Your account should look like this:
https://ams.ukbiobank.ac.uk/ams/resProjects
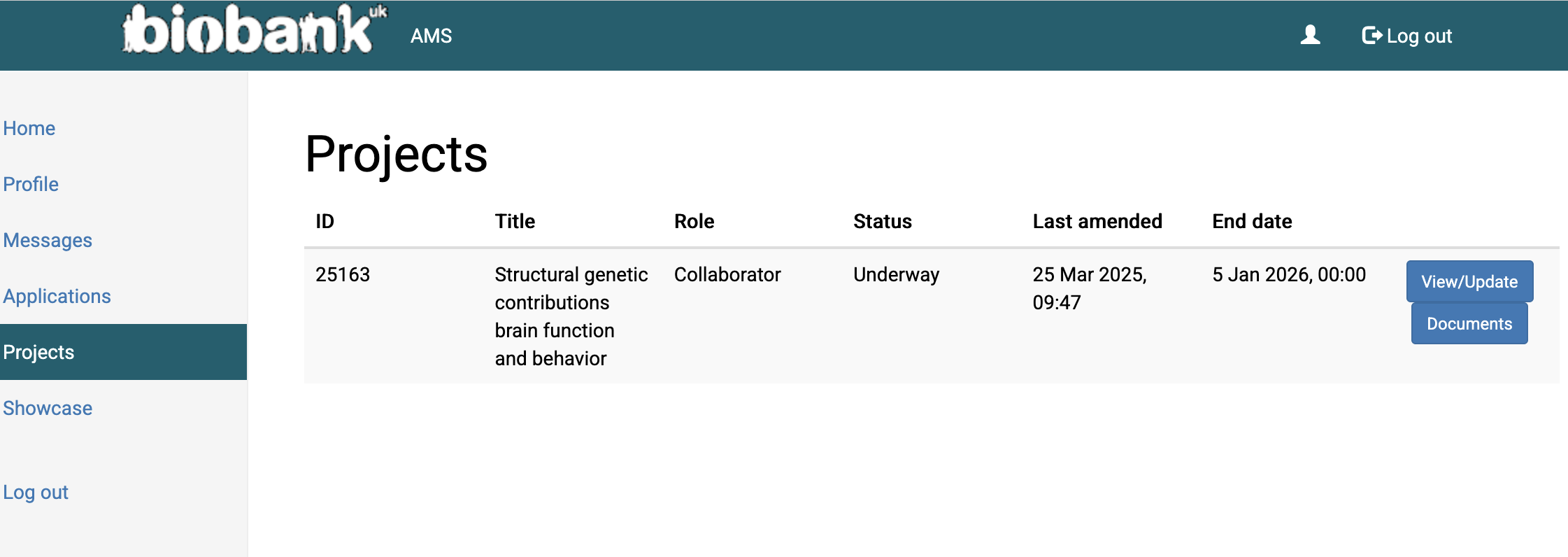
2. Do the following courses:
The courses you need to do are listed in this section within your AMS profile:
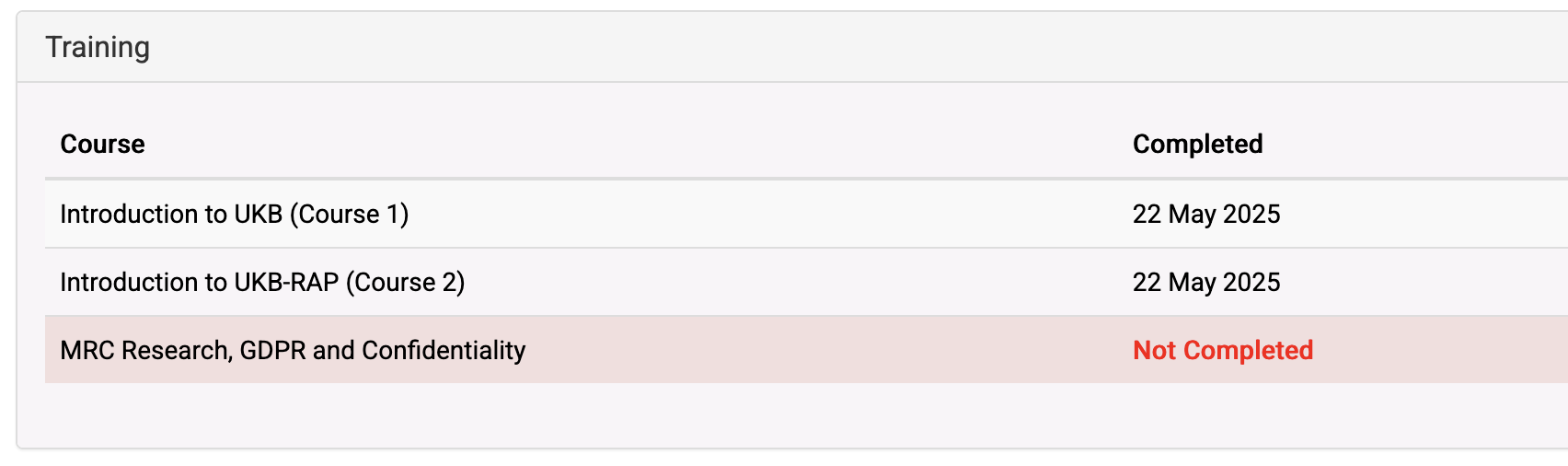
Example of courses— this person has completed all but the MRC Research Course
- UKB course 1 - on UKB Profile
- Watch modules, take quiz
- UKB course 2 - on UKB Profile
- Watch modules, take quiz
- The MRC course consists of 10 short video modules on various aspects of GDPR data security, including identifiable data, data sharing, and safeguards.
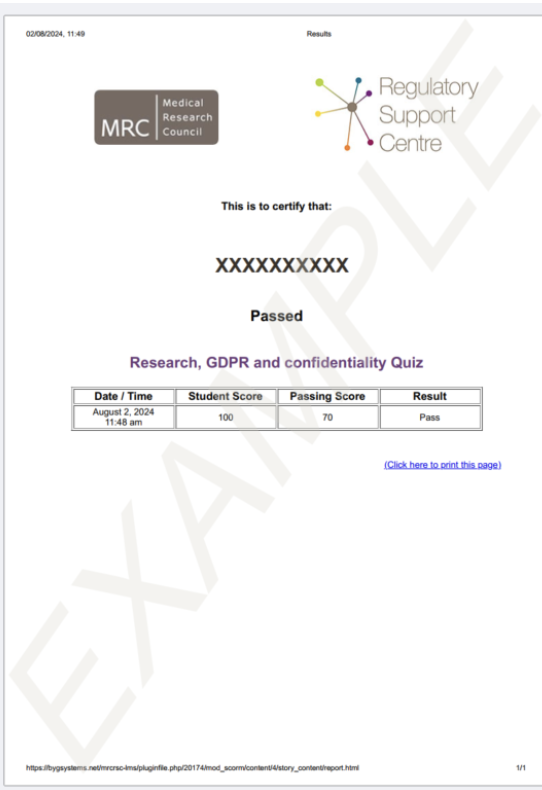
- To complete the course, you must pass a 10-question multiple-choice quiz with a score of 70% or higher. Your certificate should look like this:
- Certificate image reference
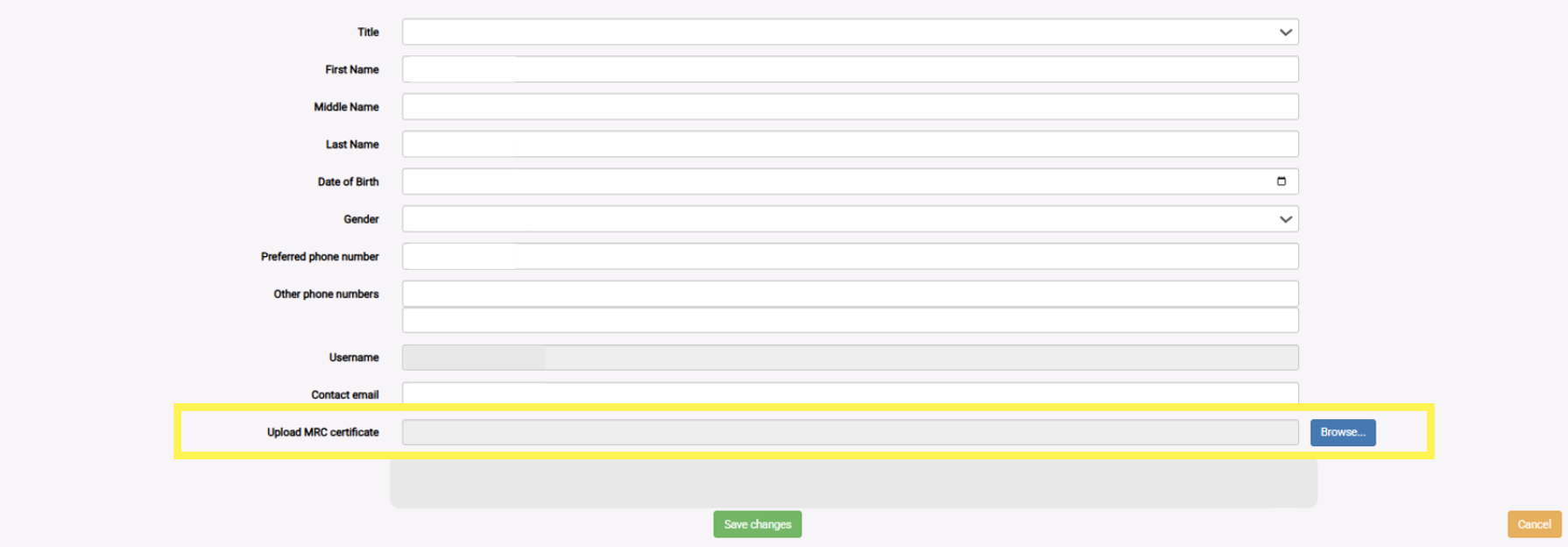
- Steps to upload your certificate:
- Log in to your AMS account
- Select ‘Profile’
- Scroll down to ‘Upload MRC certificate’
- Select the blue ‘Browse’ button to the right
- Upload your certificate in PDF format
Where to upload your certificate image reference
Listed on here: https://community.ukbiobank.ac.uk/hc/en-gb/articles/22145292393757-Researcher-Training-Courses
3. Get Approval and Dispense Data
- Once approved, you’ll get access to UKB-RAP via the DNAnexus cloud platform.
- You’ll receive a RAP project with the approved data preloaded.
4. Log In to UKB-RAP
- Use your credentials to log in at: ukbiobank.dnanexus.com
- You’ll be taken to your project workspace in the cloud.
5. Set Up Your Environment
- Choose between:
- JupyterLab: Interactive Python/R environment
- Terminal (bash): For CLI tasks
- Install additional libraries (Python, R, bash) as needed
- Use the Spark SQL database for fast querying of tabular data
To learn how to use and manipulate data in the UKB-RAP, the documentation is here
UKB-RAP Documentation
Training resources
The UKB-RAP training resources page allows you to specifically navigate to different video tutorials of the UKB-RAP. Some particular videos of note include the overview training videos. [Part 3](https://www.youtube.com/watch?v=foB7y2ZJHF4) covers the cohort browser and how to explore and extract data, part 4 talks about how to extract phenotype data using both table exporter and dx extract_dataset, whilst part 5 talks about the tools library with a focus on using Swiss Army Knife.
Additionally, a particularly notable webinar video (found here) guides you through data dispensal, project creation, data refreshing, dataset types and structures, basic file operations, apps in the UI and the considerations of cloud based analysis.
DNAnexus documentation
The DNAnexus documentation page provides detailed information on how to perform operations in the UKB-RAP using both the command line interface (CLI) and the user interface (UI), including how to run apps and workflows, as well as operate the cohort browser. A particularly essential resource for users of the CLI is the index of dx commands page, which you can use to search for commands to perform operations like [deleting](https://documentation.dnanexus.com/user/helpstrings-of-sdk-command-line-utilities#rm), uploading and downloading data. If you have questions about what data you can download, please see the following community forum post.
UK Biobank Github notebooks
To help researchers understand how to use the UKB-RAP, UK Biobank has created the UKB GitHub to offer code examples and insights for data analysis on the platform. This involves looking at how to access, extract and analyse data covering a range of data types, using Jupyter notebooks for both python and R, as well as Rstudio notebooks. More specifically, the [A-series (Accessing Data) notebooks](https://github.com/UK-Biobank/UKB-RAP-Notebooks-Access) focus on how to access and examine UKB phenotypic data, and the G-series (Genomics) notebooks focus on performing genomics analytics workflows. Additionally, there are notebooks which allow individual SNPs to be filtered from the UKB genotyping data, as well as notebooks for researchers interested in executing complex, multi-stage workflows on the UKB-RAP via apps, applets and Workflow Description Language (WDL). To learn more about these notebooks and how to access them, please see our UK Biobank GitHub notebooks article on the community forum which explains what there is on offer.
On a related note, DNAnexus also operates a a GitHub which offers some notebooks that you may find useful. For example, the basic data extraction notebook guides you on how to extract phenotypic data, and the dx data notebook can help you to load, access and retrieve metadata within datasets and cohorts.
Misc Documentation Pages:
Setting up a project in the UKB-RAP
Finding data and how it is organised
Working with Jupyter Notebooks
How to access and use dispensed data
Connect your UK Biobank AMS account to the UKB-RAP
Bulk data and using the tool library
How long does it take to be up and running with the data?
UKB-RAP costs
For guidance on how much money you can expect to spend for computation and data storage on the UKB-RAP, please see the [UKB-RAP Rate Card](https://20779781.fs1.hubspotusercontent-na1.net/hubfs/20779781/Product Team Folder/Rate Cards/BiobankResearchAnalysisPlatform_Rate Card_Current.pdf) where you will find information on the cost per hour of using different instance types. You may also want to see our costs and billing page and the costs & financial support webpage. To help reduce costs, remember to maintain good file management.
Find support
The UKB-RAP help centre provides a page which links to various UKB-RAP support resources.
If you have further questions about using the UKB-RAP, the UK Biobank community forum is a great place to browse previously asked questions and to post your own inquiries to help out the wider research community. Alternatively, you can submit a ticket to get help from UK Biobank or contact DNAnexus support at ukbiobank-support@dnanexus.com for assistance with any technical issues you may be facing.
If you are interested in looking through the metadata for fields in the UKB-RAP, you may also be interested in looking at the Showcase website. Please see this article for more details.
FAQs
Registering:
What do I need to register for access?
Verifying an email address after submitting your AMS registration
How long will it take for my registration to be reviewed?
How can I confirm my place of work?
How do I update the institute section of my registration?
Applying for Access:
Will UK Biobank complete a supplier set up form or supplier questionnaire issued by my institution?
How to: Complete an access application
Sign a Material Transfer Agreement (How to VIDEO)
Who should I provide for an MTA contact?
How do I add an MTA contact to my application?
Introductory Training:
How do I upload my MRC certificate to my AMS profile?
I’ve completed training, when will my profile be updated?
I have completed my training, so why can’t I access the UKB-RAP?
Managing Projects:
How can I remove a collaborator from my project?
How do I extend the scope of my project?
How do I change the tier of my project?
How do I extend the duration of my project?
Who should I list as the DPO contact at my institute?
How to create a UKB-RAP account
Make a change request for your project in AMS (How to VIDEO)
How do I add researchers to my project?
How do I access the restricted home location data?