fMRIPrep Overview
[PAGE IN PROGRESS]
Documentation Webpage: https://fmriprep.org/en/stable/outputs.html 1
General Overview
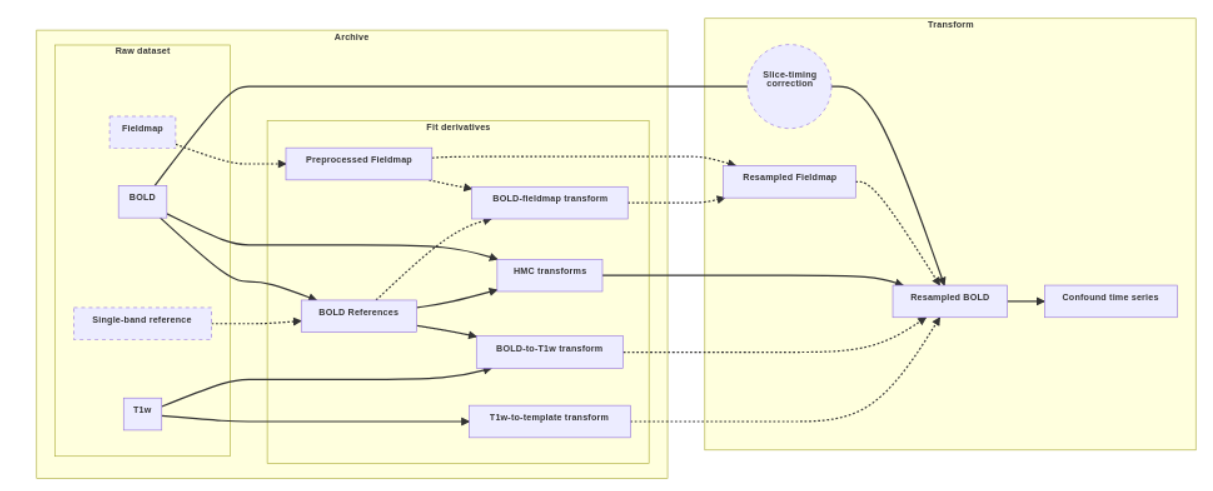
Diagram of fMRIPrep, 04-26-24 Dashes indicate optional data or processes. The “fit” derivatives include a collection of individual volumes and transform files. The “transform” section shows the process used to generate resampled BOLD series. The available inputs, such as fieldmaps and slice-timing metadata, and the target space, such as an MNI template, determine the final result.
Step 1) Anatomical preprocessing
- Brain Extraction
- Brain tissue segmentation(making brain mask)
- Spatial nomalization(e.g MNI152LinAsym:res-2)
Step 2) BOLD preprocessing
- HMC(We use reference BOLD image gained from t1-saturated image)
- STC
- SDC
- resampling BOLDs into native spaces(This part was very tricky for me. I understood this process as followings. 1. combine each transform matrix into one transform matrix 2. apply one transform matrix onto each volume images)
- EPI to T1w registration (re-align EPI images onto normalized t1w)
- finally, normalize EPI images onto standard image.
*Note: We try to resample as few times as possible. So while we do always resample to native (STC + HMC + SDC), that is used for processes that need data in native BOLD space. When we resample to T1w space, we apply (HMC + SDC + BOLD-to-T1w) in a single step from the STC’d (if applied) time series.